Neural Network on Microcontroller (NNoM)
NNoM is a higher-level layer-based static Graph Neural Network library specifically for microcontrollers.
NNoM is released under LGPL-V3.0, please check the license file for detail.
Dependencies
NNoM currently runs on top of CMSIS-NN/DSP backend.
Therefore, it runs on ARM Cortex-M 32-bit RISC processor only.
Why NNoM?
The aims of NNoM is to provide a light-weight, user-friendly and flexible interface for fast deploying.
If you would like to try more up-to-date, decent and complex structures on MCU (such as Inception, SqueezeNet, ResNet, DenseNet...)
NNoM can help you to build those complex structures in the same way as you did in Keras.
Most importantly, your implementation can be evaluated directly on MCU with NNoM.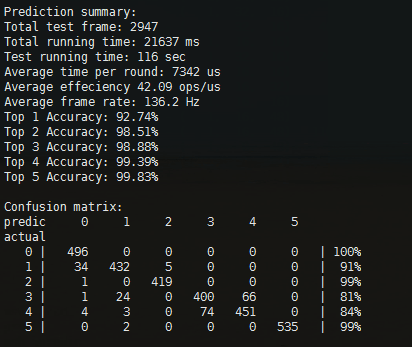
A simple example:
#define INPUT_HIGHT 1
#define INPUT_WIDTH 128
#define INPUT_CH 9
new_model(&model);
model.add(&model, Input(shape(INPUT_HIGHT, INPUT_WIDTH, INPUT_CH), qformat(7, 0), input_buf));
model.add(&model, Conv2D(16, kernel(1, 9), stride(1, 2), PADDING_SAME, &c1_w, &c1_b)); // c1_w, c1_b are weights and bias
model.add(&model, ReLU());
model.add(&model, MaxPool(kernel(1, 4), stride(1, 4), PADDING_VALID));
model.add(&model, Dense(128, &ip1_w, &ip1_b));
model.add(&model, ReLU());
model.add(&model, Dense(6, &ip2_w, &ip2_b));
model.add(&model, Softmax());
model.add(&model, Output(shape(6, 1, 1), qformat(7, 0), output_buf));
sequencial_compile(&model);
while(1){
feed_input(&input_buf)
model_run(&model);
}The NNoM interfaces are similar to Keras
It supports both sequential and functional API.
The above codes shows how a sequential model is built, compiled, and ran.
Functional Model
Functional APIs are much more flexible. An example is shown in uci-inception example
It allows developer to build complex structures in MCU, such as Inception and ResNet.
The below codes shows an Inception structures with 3 parallel subpathes.
#define INPUT_HIGHT 1
#define INPUT_WIDTH 128
#define INPUT_CH 9
nnom_layer_t *input_layer, *x, *x1, *x2, *x3;
input_layer = Input(shape(INPUT_HIGHT, INPUT_WIDTH, INPUT_CH), qformat(7, 0), input_buf);
// conv2d
x = model.hook(Conv2D(16, kernel(1, 9), stride(1, 2), PADDING_SAME, &c1_w, &c1_b), input_layer);
x = model.active(act_relu(), x);
x = model.hook(MaxPool(kernel(1, 2), stride(1, 2), PADDING_VALID), x);
// parallel Inception 1 - conv2d
x1 = model.hook(Conv2D(16, kernel(1, 5), stride(1, 1), PADDING_SAME, &c2_w, &c2_b), x); // hooked to x
x1 = model.active(act_relu(), x1);
x1 = model.hook(MaxPool(kernel(1, 2), stride(1, 2), PADDING_VALID), x1);
// parallel Inception 2 - conv2d
x2 = model.hook(Conv2D(16, kernel(1, 3), stride(1, 1), PADDING_SAME, &c3_w, &c3_b), x); // hooked to x
x2 = model.active(act_relu(), x2);
x2 = model.hook(MaxPool(kernel(1, 2), stride(1, 2), PADDING_VALID), x2);
// parallel Inception 3 - maxpool
x3 = model.hook(MaxPool(kernel(1, 2), stride(1, 2), PADDING_VALID), x); // hooked to x
// concatenate 3 parallel.
x = model.mergex(Concat(-1), 3, x1, x2, x3); // new merge API.
// flatten & dense
x = model.hook(Flatten(), x);
x = model.hook(Dense(128, &ip1_w, &ip1_b), x);
x = model.active(act_relu(), x);
x = model.hook(Dense(6, &ip2_w, &ip2_b), x);
x = model.hook(Softmax(), x);
x = model.hook(Output(shape(6,1,1), qformat(7, 0), output_buf), x);
// compile and check
model_compile(&model, input_layer, x);
while(1){
feed_input(&input_buf)
model_run(&model);
}Please check A brief manual
Detail documentation comes later.
Available Operations
Layers
| Layers | Status | Layer API | Comments |
|---|---|---|---|
| Convolution | Beta | Conv2D() | Support 1/2D |
| Depthwise Conv | Beta | DW_Conv2D() | Support 1/2D |
| Fully-connected | Beta | Dense() | |
| Lambda | Alpha | Lambda() | single input / single output anonymous operation |
| Input/Output | Beta | Input()/Output() | |
| Recurrent NN | Under Dev. | RNN() | Under Developpment |
| Simple RNN | Under Dev. | SimpleCell() | Under Developpment |
| Gated Recurrent Network (GRU) | Under Dev. | GRUCell() | Under Developpment |
| Flatten | Beta | Flatten() | |
| SoftMax | Beta | SoftMax() | |
| Activation | Beta | Activation() | A layer instance for activation |
Activations
Activation can be used by itself as layer, or can be attached to the previous layer as "actail" to reduce memory cost.
| Actrivation | Status | Layer API | Activation API | Comments |
|---|---|---|---|---|
| ReLU | Beta | ReLU() | act_relu() | |
| TanH | Beta | TanH() | act_tanh() | |
| Sigmoid | Beta | Sigmoid() | act_sigmoid() | |
Pooling Layers
| Pooling | Status | Layer API | Comments |
|---|---|---|---|
| Max Pooling | Beta | MaxPool() | Support 1/2D |
| Average Pooling | Beta | AvgPool() | Support 1/2D |
Matrix Operations Layers
| Matrix | Status | Layer API | Comments |
|---|---|---|---|
| Multiple | Beta | Mult() | |
| Addition | Beta | Add() | |
| Substraction | Beta | Sub() | |
| Dot | Under Dev. | ||
Memory requirements
NNoM requires dynamic memory allocating during model building and compiling.
No memory allocating in running the model.
RAM requirement is about 100 to 150 bytes per layer for NNoM instance, plus the maximum data buf cost.
The sequential exmaple above includes 9 layer instances. So, the memory cost for instances is 130 x 9 = 1170 Bytes.
The maximum data buffer is in the convolutional layer.
It costs 1 x 128 x 9 = 1152 Bytes as input, 1 x 64 x 16 = 1024 Bytes as output, and 576 Bytes as intermedium buffer (img2col).
The total memory cost of the model is around 1170 (instance) + (1152+1024+576)(network) = ~3922 Bytes.
In NNoM, we dont analysis memory cost manually like above.
Memory analysis will be printed when compiling the model.
Deploying Keras model to NNoM
No, there is no single script to convert a pre-trained model to NNoM.
However, NNoM provides simple python scripts to help developers train, quantise and deploy a keras model to NNoM.
Please check A brief manual
and UCI HAR example.
The tutorial comes later.
Porting
It is required to include the CMSIS-NN lib in your projects.
The porting is easy on ARM-Cortex-M microcontroller.
Simply modify the nnom_port.h refer to the example in the file.
Current Critical Limitations
- Support 8-bit quantisation only.
- Support only one Q-format in one model.
- Cannot free the memory allocated by model.
TODO
- Support RNN types layers.
- Support mutiple Q-formats
- support memory releasing.
Contacts
Jianjia Ma
J.Ma2@lboro.ac.uk or majianjia@live.com
Citation Required
Please contact us using above details.
